Biography
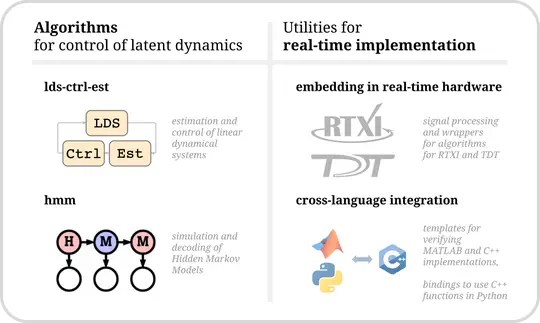
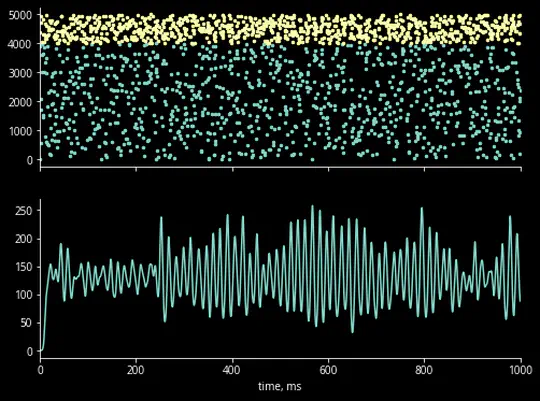
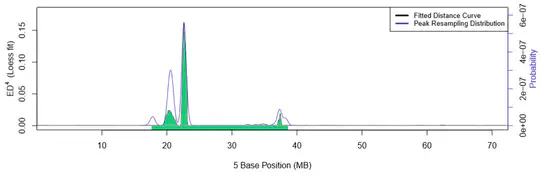
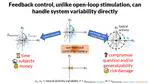
Hello there! I’m a PhD student advised by Chris Rozell at Georgia Tech, developing advanced closed-loop optogenetic control techniques for neuroscience. Specifically, I am applying optimal, model-based control to manipulate population-level variables of neural activity.
I’m fascinated by the biological computational principles and how we can exploit them for next-generation AI/ML technologies. And vice-versa: how we can apply modern ML to neurotech/neuroscience, especially for building life- and time-saving brain-computer interfaces. I plan on looking for an industry research scientist role post-graduation.
Download my CV (last updated 2023).
- NeuroAI: Bridging theoretical neuroscience and machine learning
- Brain-computer interfaces
PhD in Biomedical Engineering, in progress
Georgia Institute of Technology/Emory University
BS in Bioinformatics, 2019
Brigham Young University
Projects
Recent Posts
Publications
Contact
- kjohnsen@gatech.edu
- 756 W Peachtree St, S1110C, Atlanta, GA 30308
- DM Me